Nvidia Buys Illumina? The AI Foundry for Healthcare – The Hardware of Life
Overcoming Eroom's Law: Enabling Moore's Law in Healthcare With Potential Acquisition
All of you are familiar with Moore’s Law, the observation that the number of transistors on a microchip doubles every two years. We doubt many of you are familiar with the cousin of Moore’s Law, Eroom’s Law.
Yes. Eroom’s Law is literally just Moore’s Law spelled backwards.
No. We didn’t make it up, but we wish we did!
Eroom’s Law, in an ironic twist, revolves around the concept that the cost of developing a new drug tends to double approximately every nine years.

Moore's Law and Eroom's Law both examine three common factors in the fields of Semiconductors and Drug Research: time, money, and the unit of output.
Semiconductors, the backbone of all technological progress in the modern era, have improved exponentially in unit cost and capabilities thanks to Moore’s law. Drug discovery, one of the largest areas of spending in healthcare has seen unit costs expand tremendously, requiring billions of dollars to bring a new innovation to the market.
While many argue about the death of Moore’s law in the semiconductor industry, the spirit of Eroom’s Law is clearly alive and well in healthcare.
Transcending Eroom’s law, and putting improvement in healthcare back on track with the rest of humanities technological progress will require humanity to unleash the existing advances of cloud, accelerated compute, and AI on the hardware of life.
Healthcare is the largest area of the economy, more than 4 trillion dollars in the US alone. It continues to be largely ignored by big tech and the massive technological progress delivered by Moore’s Law.
Eroom’s Law Across Healthcare Verticals
Though the pharmaceutical industry has taken several steps to improve the drug discovery process and probability of success for new therapeutic modalities, the overall ratio of research and development cost to revenue in the pharmaceutical industry continues to creep higher. This trend has continued despite growth in healthcare costs (and also pharmaceutical company revenues) well above the consumer price index (CPI) average. This is largely because predicting if a drug will make it through the incredibly lengthy and costly clinicals is difficult due to our lack of understanding of biology and various interactions between systems.
A large reason for the diminishing return on investment in recent years is the fact that the drug discovery and research process has yet to fully leverage the potential of technological advancements, notably the use of cloud computing and artificial intelligence.
An empirical manifestation of Eroom’s law can be seen in the fact that healthcare outcomes have seen diminishing returns on investment, with life expectancy in the US only increasing from 70 years to 79 years from 1960 to 2019 despite per capita healthcare expenditure increasing from ~5% to ~20% of the US GDP. The 21st century has seen costs get exponentially worse and therefore less accessible in the developed world.
Many areas in healthcare have seen well below their potential in terms of improvements. From medical records to the ubiquity of the Apple watch and Peloton bike, there has been an exponential growth in data generated and scientific interest in harnessing said data, but little to show in terms of results. We are again missing key analytical infrastructure to process the data and decipher signal from noise in order to provide real-time insights into patient medical history, biological signals (heart rate, oxygen and glucose levels), and environmental impacts on patient outcomes.
In the field of medical records, despite electronic health record (EHR) adoption in hospitals rising from 10% in 2008 to 95% by 2014 there has been little progress on extracting insights from data, either at a single patient level or across large sets of patients. Electronic health records hold structured data such as the patient demographics, medication, diagnostic data (labs, imaging, microbiology, pathology, etc) and immunizations.
However most data generated in an EHR is unstructured data such as physician notes, insurance company data, and billing data. Most of the immense volume of healthcare data being generated, estimated to have reached 2,314 exabytes of data by 2020, goes unharnessed.

For instance, Emergency Room doctors have, at best 5-10 minutes, and at worst seconds in the most acute cases, to pore through this unstructured data in the form of physician notes and scans that can span hundreds of pages. With many hospitals using different electronic health record (EHR) implementations, the inability to easily access EHRs for a patient from a different healthcare provider or run data analysis across a huge number of EHR records robs the healthcare industry of access to game changing data to drive drug development and learnings for better clinical treatment.
Using Large Language Models (LLMs) to ingest the huge volumes of unstructured data within electronic health records could enable the use of this information in diagnosis or treatment, particularly in urgent and critical care, and provide a wealth of insights in research settings. LLMs are also well suited towards categorizing data and streamlining data portability issues, potentially addressing the current situation where individual patients’ EHRs at different hospitals or clinics effectively live on their own island.
Our friend Gaurav at GettingClinical expands on this idea in much greater detail here. In future reports, we will be diving into LLMs for healthcare as well.
Flipping The Script: From Eroom’s Law Back To Moore’s Law
While healthcare costs have increased, there are fields within the healthcare industry that have in fact exhibited better than Moore’s Law scaling. The first human genome was sequenced in 2003, cost $3B, and took 13 years of work by multiple institutions. By 2008, the cost of sequencing a whole genome was $2M, by 2014 this fell to $1,000 and is quoted at $200-$600 today.

The huge drop in cost per sequencing has led to an exponential growth in genomics data. This improvement isn’t slowing either, Illumina, the leading genome sequencing tool company, recently released the NovaSeq X instrument in September 2022 which triggers another shrink in upstream sequencing costs. The cost per sequence for this tool is ~$200, an 80% reduction compared to the prior generation tool. Despite this reduction, the time to sequence still remains a key bottleneck in clinical adoption with the newest instrument still requiring 21-48 hours of run time for sequencing.
Between 2018 (~100 Petabytes) and 2021 (280 PB), data created on instruments sold by Illumina, the leading gene sequencing machine company, exhibited a Compound Annual Growth Rate (CAGR) of 41%, a doubling every 2 years. This trend is only accelerating.
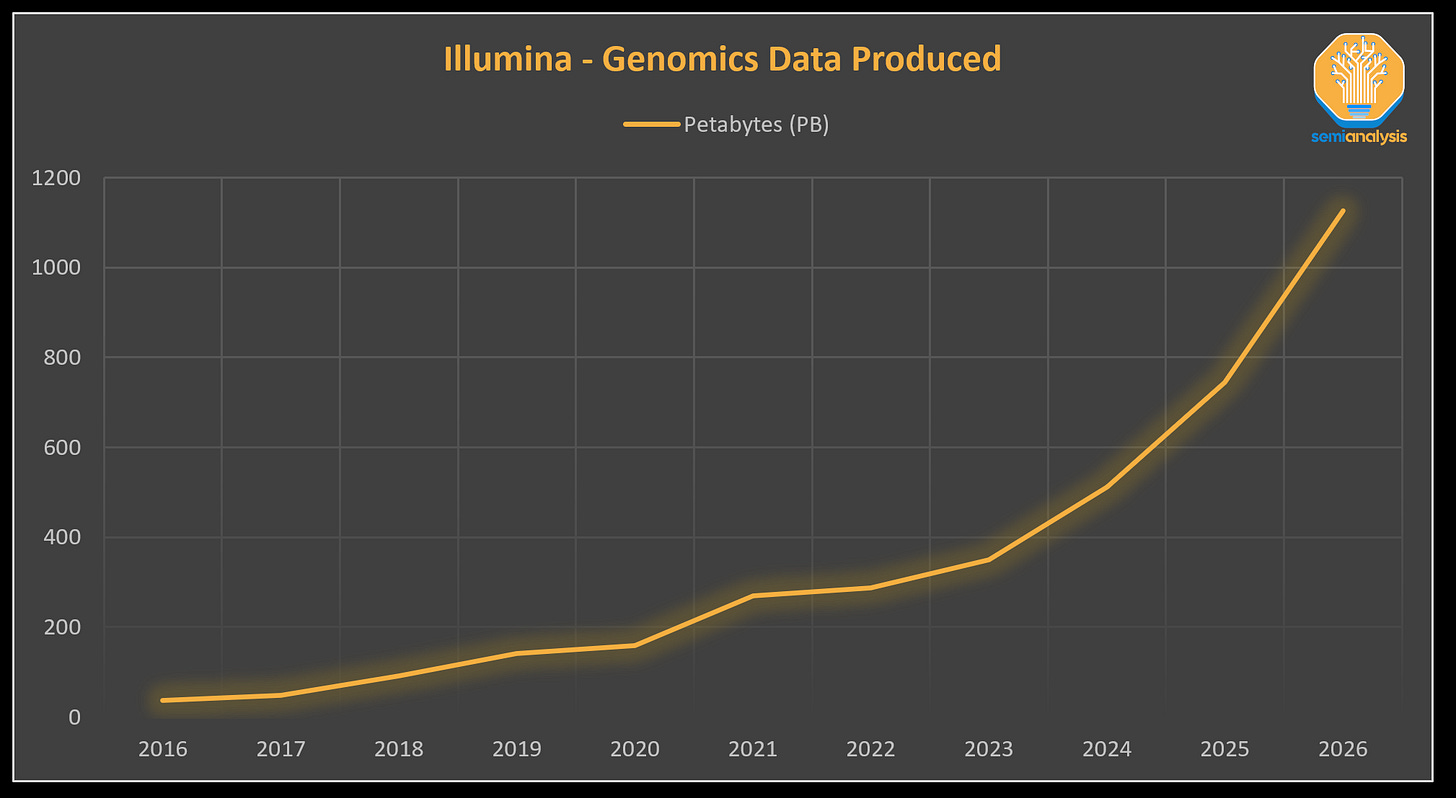
The volume of data generated from genomics projects is immense, and is estimated to require up to 40 Exabytes of annual storage needs by 2025, an order of magnitude larger than annual storage needs estimated at YouTube in 2025 and a volume of data that is 8x larger than would be needed to store every word spoken in history.
If we consider Data as the new Oil, and the potential for large language models to disrupt numerous industries, then genomics becomes one of the most crucial areas to initiate this transformation. Within the field of genomics, over 90% of the generated data is produced by Illumina sequencers.
This proliferation of genomics data has the potential to unlock applications in disease diagnosis, personalized medicine, screening and prevention, cures for intractable diseases, cancer treatment, and in critical care settings. It should be noted that genomic data is not yet directly related to drug discovery and Eroom’s law, but it is a factor, and genomics has already began bending the curve with advancements such as DECLs. Furthermore, time to market is the largest cost in drug development, and genomics progress will help accelerate cost improvements.

Just as AI required Moore’s Law to drive down the cost of compute low enough to enable image nets then eventually language models, a high unit cost of sequencing a human genome is no longer the hurdle limiting advancements as it once was in the past. Today, less than 10% of the total cost of sequencing the human genome is related to the actual act of sequencing DNA. More than 90% of the cost is associated with gathering samples, data management, data reduction, and secondary analysis.
The missing piece of the puzzle is in further developing the data analysis infrastructure and harnessing accelerated compute capabilities needed to bring various research into widespread clinical use. The lack of easy to use platforms and tools is one of the chief reasons adoption is so low.
For example, Oncology is a whopping $78B Genomics opportunity, yet penetration of genomics use within this field is only ~2%.

Further adoption of whole genome sequencing opens a pandora’s box. The more patients that are sequenced for diagnostic or research purposes, the more researchers can associate genetic mutations with diseases and develop treatments, building a more comprehensive test and treatment dictionary. The more useful the sequencing process is to patients in a clinical or diagnostic setting, the more demand for the use of genomics will increase.
However, the more we learn about genomics and the more treatments we develop to address those genetic mutations, the more questions we are asking. Genomics only provides one type of insight.
More on other “Omics”, Pandora’s Box
From the human genome to metabolic pathways, scientists are gleaning a deeper and deeper understanding of the human body. The data we are gathering about previously disparate disciplines such as genomics and proteomics are increasingly intertwined.
Proteomics, or the study of the structure and functions of proteins, is another important field to follow. The structure of proteins can meaningfully change when binding with other molecules in the body. This is essential to determining targets for therapies or understanding how cells respond to previous therapies. Prior to the advent of accelerated computing, protein structure analysis was an incredibly complex, laborious, and time-consuming task.
Look no further than the groundbreaking impact DeepMind’s AlphaFold has had on this field. The work at DeepMind has solved the 50 year old problem of protein folding in biology to an unprecedented accuracy of less than 0.1 nanometers. These advances have not only solved several old problems but have also illuminated a number of new opportunities. For example, AlphaFold’s successors are now helping researchers better understand how and where to deliver therapies to maximize clinical utility.
Epigenomics explores how genes can be expressed differently due to changes resulting from chemical compounds or proteins that attach to DNA as opposed to changes in the actual DNA sequence itself. One commonly studied example is DNA methylation, where a methyl group (-CH3) is attached to the cytosine base, causing a cell to recognize this tag and treat the gene differently. These changes can result from environmental/external factors and can themselves cause diseases.
Historically these disparate disciplines (the various “omics”) were explored in their own silos.
Subsequent reports will dive into the advances in each of briefly mentioned fields as well as others not mentioned, especially with regards to AI usage and demand.
The healthcare sector finds itself on the precipice of a paradigm shift as it starts to embrace these key enabling trends and continues to unify the various fields of research. Recent advancements in a host of fields from Genomics to Gene Therapies, Drug Development and a proliferation of electronic health care data have outpaced the industry’s ability to fully harness these transformative innovations. Instead of seeing an acceleration in breakthroughs, the industry has hit a wall. There is a clear potential to dismantle Eroom's Law.
With the advent of cloud computing, massively scaled parallelized accelerated compute as well as the development of various machine learning / large language models, humanity finally has the tools to tear down these roadblocks for each vertical of healthcare. Overcoming these obstacles will lead to a massive boom in demand for accelerated compute / data analysis and create a huge revenue opportunity for the company that can be the AI foundry for these applications.
An AI foundry could tap the >$4T US market for healthcare and partner with the industry to consolidate R&D efforts into a standardized, easy to use stack, implementing easily accessible and affordable cloud-agnostic/on-premises capable analytics, and as a result create economies of scale across the entire ecosystem to drive healthcare costs down.
Before we dive into the specific synergies and opportunities related to Nvidia acquiring Illumina, let’s go through a brief overview of genomics. Our friends from Asianometry have provided a detailed overview of the history around next generation sequencing, explaining how Illumina is the ASML of genomics.
Genomics: Turning Analog Samples into Digital DNA
Genomics is the study of the entirety of an organism’s DNA sequence (its “genome”). DNA is the densest form of data in the world, and it makes modern semiconductors such as 3D NAND look like child’s play. The 3.2 billion base pairs that form a human’s DNA contain ~1.5GB of data encoded into a molecule chain tightly wound up to fit in a space less than 0.0002 inches wide.
Samples come in many shapes, sizes, and states. In their existing form our ability to interrogate them and draw insights is limited. Blood is red. Saliva is wet. Tissues are squishy. We like to think of gene samples as “analog samples” and sequencing as a form of analog to digital conversion. Genomics enables a host of research applications from AlphaFold to gene therapies.
Of the 3.2B base pairs in the human genome, only 2% actually contain the approximately 20,000 genes that form the instructions to construct amino acids and thus proteins that are vital to carrying out critical bodily functions and cellular processes. The other 98% of the DNA sequence is referred to as “non-coding” DNA.
Previously it was thought this non-coding DNA had no function, but scientists now believe that this large part of the genome is still vital towards influencing whether and how genes are activated. Study of non-coding DNA has also accelerated research around epigenomics, the study of how modifications that don’t directly change the DNA sequence can nonetheless affect gene activity, among other fields.
Making a Long Story, Short: Short and Long-Read Sequencing
There are two different approaches to DNA sequencing: short-read and long-read.
Short-read sequencing, as the name suggests, focuses on examining a small segment of the genome at a time by breaking up the entire genome into many fragments of between 50 to 300 base pairs in length. Long-read sequencing, as the name suggests, provides a more complete picture of the genome with less ambiguity, scanning 5,000 to 30,000 base pairs per fragment.
Choosing between the two modalities comes down to the specific goals users are trying to accomplish. Short-read sequencing dominates the market due to higher accuracy, lower error rates, faster sequencing times, and cheaper costs. Long-read sequencing is helpful for studying complex regions of the genome, particularly for regions that contain repetitive sequences of base pairs, making alignment of these regions less ambiguous.
The market is generally dominated by Illumina who focuses on short-read machines. The UK Competition and Market Authority (CMA) estimates Illumina has 80-90% market share of the worldwide sequencing market.
The market for Long-Read sequencing machines is slightly more fragmented with firms such as Pacific BioSciences (PacBio) and Oxford Nanopore. Upstarts in the short-read market include Element Biosciences, Singular Genomics, and Ultima Genomics while industry giant Thermo Fisher participates through their Ion Torrent platform.
Subsequent reports will dive deeper into the short vs long-read market and the competitive dynamics.
Though each platform possesses its own strengths and weaknesses, the underlying processes and workflows follow the same path. Two key bottlenecks in the process revolve around the interoperability of data during base calling and alignment as well as variant calling.
Bottlenecks Building: Base Calling, Alignment, and Variant Calling
As the cost of sequencing diminishes, the prominence of compute expenses within our overall costs increases.
Al Maynard, associate director of software engineering at Illumina
The quote above shows remarkable foresight in the face of declining Human Genome sequencing costs that outpace Moore's Law. This, combined with demand's price elasticity, results in a data production surge due to the cost of Human Genome sequencing costing only $200 now. However, these gains now face Eroom's Law as sequencers generate raw data. Scientists rely on extensive computational processes for contextual insight and analysis, known as secondary or downstream analysis. The cost and time of secondary analysis far outstrips sequencing, representing future opportunities for Nvidia to flip the script on Eroom’s Law.
Base calling, the initial step in sequencing data, is the process by which the individual letters of the DNA sequence (A,T,G or C) are determined by interpreting fluorescent or electrical signals (depending on the technology used) generated from the sample DNA. However, there are significant sources of error in processing these signals. Base calling accuracy plays a vital role in determining the quality of all subsequent steps in secondary analysis. As the saying goes, "garbage in, garbage out." An effective base caller must correct for signal error and noise and accurately output the letters of the sequence that are in the genome. A deep neural network is well suited to base calling as it can be trained to approximate almost any function and correct for such errors and noise, but implementing this approach requires significant compute resources.
Alignment is the process of re-assembling the fragments of DNA scanned and determining where on the genome these fragments originated from, essentially reconstructing a whole picture of the original genome from these hundreds of thousands of puzzle pieces.
After reconstructing the whole genome sequence for the sample, we can proceed to variant calling, the next phase of the sequencing process. During variant calling, we detect differences or "variances" between the sequenced DNA and a reference genome. These variances allow us to identify mutations with potential functional impact. This process requires a large amount of memory and parallelization as the reads from the sequence, the reference genome, and subsequent discrepancies are simultaneously processed.
Nvidia and PacBio: Benefits of Going Full-Stack in Long Reads
PacBio, a prominent manufacturer of long-read sequencing machines, recently introduced a new instrument, the Revio, integrating Nvidia’s GPUs. Here is PacBio explaining all the benefits they realized through their partnership with Nvidia:
By using advanced GPUs from Nvidia, our team began moving workloads to GPU, starting with the GPU polish step, which had consumed over 80% of the CPU time on the Sequel IIe system. … our team was able to move additional compute steps to GPU, and was ultimately able to deliver an implementation of CCS (circular consensus sequencing) that could complete its task in 2.5 hours, generating considerable headroom for us to do even more with the newly available onboard compute afforded by adding GPU.
Integrating Nvidia’s GPUs flipped the script for PacBio. Compared to prior generations of instruments, the Revio produces 12x the amount of data in 20% less time (15x faster). To provide context, the NovaSeq X, the new instrument from Illumina highlighted earlier, relies on CPUs to produce 2.5x the amount of data compared to prior generations (16Tb vs. 6Tb), but it requires 10% more time (48 vs. 44 hours).

It's highly improbable that consumable costs in the upstream process will decrease as significantly as they have over the past 20 years, having plummeted from $3,000,000,000 to $200, instead, the bottleneck for realizing clinical adoption is shifting to secondary analysis.
The Intensive Care Unit Goes Full-Stack
What does genomics look like in a clinical setting? How can it save lives?
In late 2020, researchers from Stanford University School of Medicine conducted ultra-rapid whole genome sequencing on patients in intensive care to diagnose rare genetic diseases in hours setting a world record.
Central to this advancement was integrating machine-based and cloud-based compute. The workflow took raw signals from the instrument and transmitted them to cloud storage in real time. From there, data was distributed across multiple cloud computing instances for near real time base calling and alignment. Researchers reduced base calling and alignment time from 18.5 hours using machine-only compute to 2 hours via parallelization. The workflow used 16 instances, consisting of 48 CPUs and 4x Tesla V100s for each instance instead of the 4x Tesla V100s onboard the PromethION 48 (Oxford Nanopore).
In one case, a thirteen-year-old boy was brought to Stanford Hospital with symptoms of rapidly progressing heart failure. There were two possible causes, either an irreversible genetic heart defect requiring heart transplantation or an often self-limiting and treatable inflammatory condition known as myocarditis. The team utilized this ultrarapid sequencing process to achieve a sequencing time of 11.25 hours, beating the prior world record by 8 hours and standard of care testing by almost 2 months. They were able to determine the presence of a cardiac genetic defect and the patient subsequently underwent successful heart transplantation, ultimately saving his life. In this case, timely diagnosis was vital for appropriate and definitive treatment, highlighting just one of the many potential uses of this technology.
The per patient cost of conducting the rapid sequencing procedure was calculated to range from approximately $4,900 to $7,300 of which the cost of compute was $568 per patient on average, with the compute cost portion almost entirely driven by base calling, alignment and variant calling.
Note this is using 6-year-old V100s at inflated pricing. Costs are much lower today for these same compute resources. The authors stressed that the cost of the test is well below the $10,000 daily cost of care in an intensive care unit, and thus the test procedure is likely to be economical at scale and eventually eligible for reimbursement by insurance.
Computational Bottlenecks in Genomic Sequencing: GPUs to the Rescue
Nvidia is addressing computational bottlenecks in base calling, alignment, and variant calling through Parabricks, their genomics workflow, which is part of Clara, their suite of AI-accelerated solutions for the healthcare ecosystem.
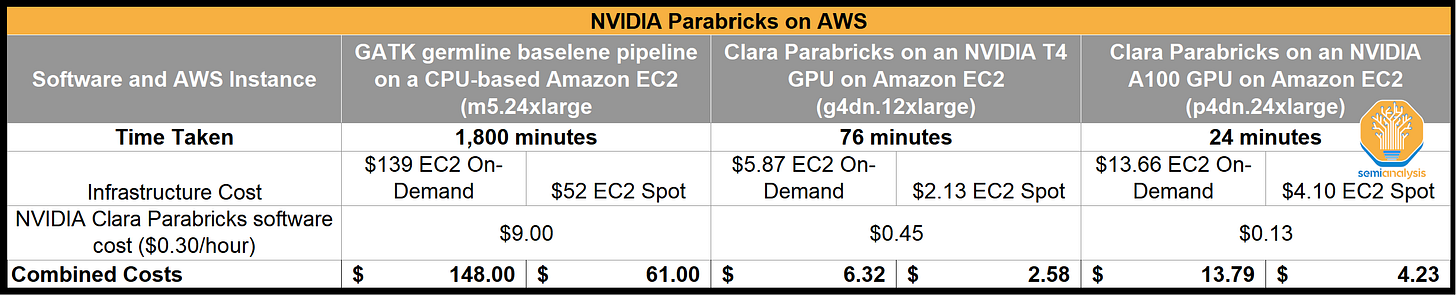
A study completed by AWS and Nvidia provides a detailed look at CPU-based analysis versus GPU-based analysis. What took 1,800 minutes of compute on CPU-based Amazon EC2, took 76 minutes on Nvidia T4 GPU, and 25 minutes on Nvidia A100 GPU. Total cost was reduced by 90-95% when running on GPU-based EC2.
Placing the above data within the framework of Illumina's integration of CPUs into their secondary analysis solution, DRAGEN, one cannot help but contemplate whether they might be missing the bigger picture.
The field of healthcare at large, and genomics in particular, is increasingly becoming a computational challenge. To overcome Eroom's law, we require comprehensive solutions that break down existing silos and construct an open ecosystem, harnessing state-of-the-art expertise in biology, chemistry, computation, engineering, and AI.
Nvidia’s Healthcare Push - A Front Row Seat
Nvidia has pushed into the healthcare market by being an enabler of accelerated compute at the instrument and device level and by providing biology and bioinformatics platforms that feature AI-defined software solutions as well as access to best-in-class implementation of these solutions on cutting edge GPU hardware.
And so that's why I really describe Nvidia not as a chip company, but we are an accelerated data center company. And so to compete at that level is very, very difficult. And the software investment that we've made and our ecosystem partners have made is really what differentiates us and allows us to remain -- allows us to innovate continuously at the speed of light, because we're a full-stack company.
Nvidia at JP Morgan Healthcare Conference 1/12/23
Nvidia’s approach to addressing this vast data analytics and compute need is multifaceted:
Integrating GPUs into Devices
Long-Read Sequencers. Both Oxford Nanopore and PacBio integrate Nvidia GPU’s into their instruments reducing sequencing time and enabling greater data extraction.
Nvidia IGX. IGX delivers secure, low-latency AI inference to address the clinical demand for instantaneous insights from a range of instruments and sensors for medical procedures, such as robotic-assisted surgery and patient monitoring.
Supercomputers. The Cambridge-1 supercomputer is the most powerful computer in the United Kingdom and was purposefully built by Nvidia to accelerate digital biology. AstraZeneca collaborated with Nvidia to build transformer-based neural network architectures capable of leveraging massive datasets for chemical structures. Oxford Nanopore utilizes Cambridge-1 to carry out algorithm improvements in hours, versus days. GSK, UK NHS, Peptone, and Alchemab has also had various breakthroughs in healthcare related applications by utilizing this supercomputer.
Nvidia Clara provides a suite of AI-accelerated solutions across the healthcare ecosystem.
Parabricks for Genomics. Parabricks, acquired in 2019, offers a suite of GPU-accelerated tools for secondary analysis in the genomics sequencing flow. It performs tasks like sequence alignment and assembly, which help determine the locations of genetic variations and mutations within the sequenced DNA. This is significantly faster and cheaper than using CPUs.
Nvidia Holoscan and MONAI for Medical Imaging
Holoscan and MONAI are both AI computing platforms for medical devices and medical imaging combining hardware systems for low-latency sensors and network connectivity, AI optimized libraries for inferences, and software-defined architectures for real-time processing of data coming from the devices or in the cloud.
Nvidia’s BioNeMo LLM, launched in March 2023, applies large language models (LLMs) to the study of chemistry, proteins, RNA and DNA, allowing developers to effectively train and deploy biology LLMs with billions of parameters, leading to insights that researchers can connect to biological properties or functions and even human health conditions. Products offered as part of BioNeMo include:
AlphaFold2: A deep learning model that reduces the time it takes to determine a protein’s structure from years to minutes or even seconds, just by using its amino acid sequence, developed by DeepMind and already used by over a million researchers.
DiffDock: To help researchers understand how a drug molecule will bind with a target protein, this model predicts the 3D orientation and docking interaction of small molecules with high accuracy and computational efficiency.
ESMFold: This protein structure prediction model, using Meta AI’s ESM2 protein language model, can estimate the 3D structure of a protein based on a single amino acid sequence, without requiring examples of several similar sequences.
ESM2: This protein language model is used for inferring machine representations of proteins which are useful for downstream tasks such as protein structure prediction, property prediction and molecular docking.
MoFlow: Used for molecular optimization and small molecule generation, this generative chemistry model creates molecules from scratch, coming up with diverse chemical structures for potential therapeutics.
ProtGPT-2: This language model generates novel protein sequences to help researchers design proteins with unique structures, properties and functions.
These platforms go a long way to enabling various applications, but there are also many haps in Nvidia’s current offerings for them to realize their ambitions of being a full-stack provider in healthcare when other ecosystems control all the data processed on their platforms.
Illumina = Missing Puzzle Piece
So genomics is a modality that is delivering great value to health care as well as research and drug discovery. It is the largest data generator in health care and growing rapidly. We're witnessing the continued decline in cost, enabling large-scale genomics programs to transpire. However, we need to be sensitive to the fact that a lot of times, when you advertise the cost of sequencing, it's just the cost of sequencing and not the downstream analysis, which is ultimately the insights that we need to care for our patients and to deliver insights in drug discovery … With more sequencing platforms and modalities entering the market, we are going to be pushing these 40 exabytes of genomic data out there into the world.
Nvidia at JP Morgan Healthcare Conference 1/12/23
Nvidia's acquisition of Illumina completes the puzzle in their healthcare push, linking upstream genomics data with downstream AI-driven analysis to create a new industry standard for healthcare.
We think an acquisition of Illumina answers the following critical questions:
How can you develop full-stack solutions in genomics when 90% of data sequenced is captured with only the aid CPUs, gating new data production?
How can you develop full-stack solutions in genomics when the default flow of data from Illumina tools is straight to Amazon’s cloud, a quasi-monopoly capturing incredibly high margins, but providing very weak analytical tools?
How can you create intuitive workflows democratizing healthcare when data and analytical tools reside in silos?
Who will unify and standardize these disparate data sources and workflows in an industry inherently inept at building software?
How can you flip script from Eroom’s to Moore’s Law without building AI-informed solutions capable of spanning the healthcare ecosystem?
How can you claim to be the purveyor of accelerated compute without enabling this compute for the largest and most important dataset in humanity, the hardware of life?
We already discussed how integrating GPUs into sequencers would deliver meaningful improvements in time to sequence and data generation for Illumina, who is behind competitors in this respect. Of course, integrating GPUs into their tools is low hanging fruit, only scratching the surface of why Illumina represents such a transformative acquisition not just for Illumina and Nvidia, but also humanity.
The real value of an acquisition of Illumina is because it represents the missing piece to complete Nvidia’s full-stack solution puzzle. Below for subscribers, let’s dive into the true reasons, and discuss Amazon Web Services Health Omics, cloud lock-in, multi-cloud strategies, the need for a neutral arms dealer, Nvidia DGX Cloud, Nvidia’s software as a service business model, and the competition to become the world’s AI foundry.
Furthermore, we will also share some governance and capital market related reasons that point to Nvidia and Illumina potentially preparing for this acquisition attempt, below.